Workflow Type: Galaxy
Frozen
Work-in-progress
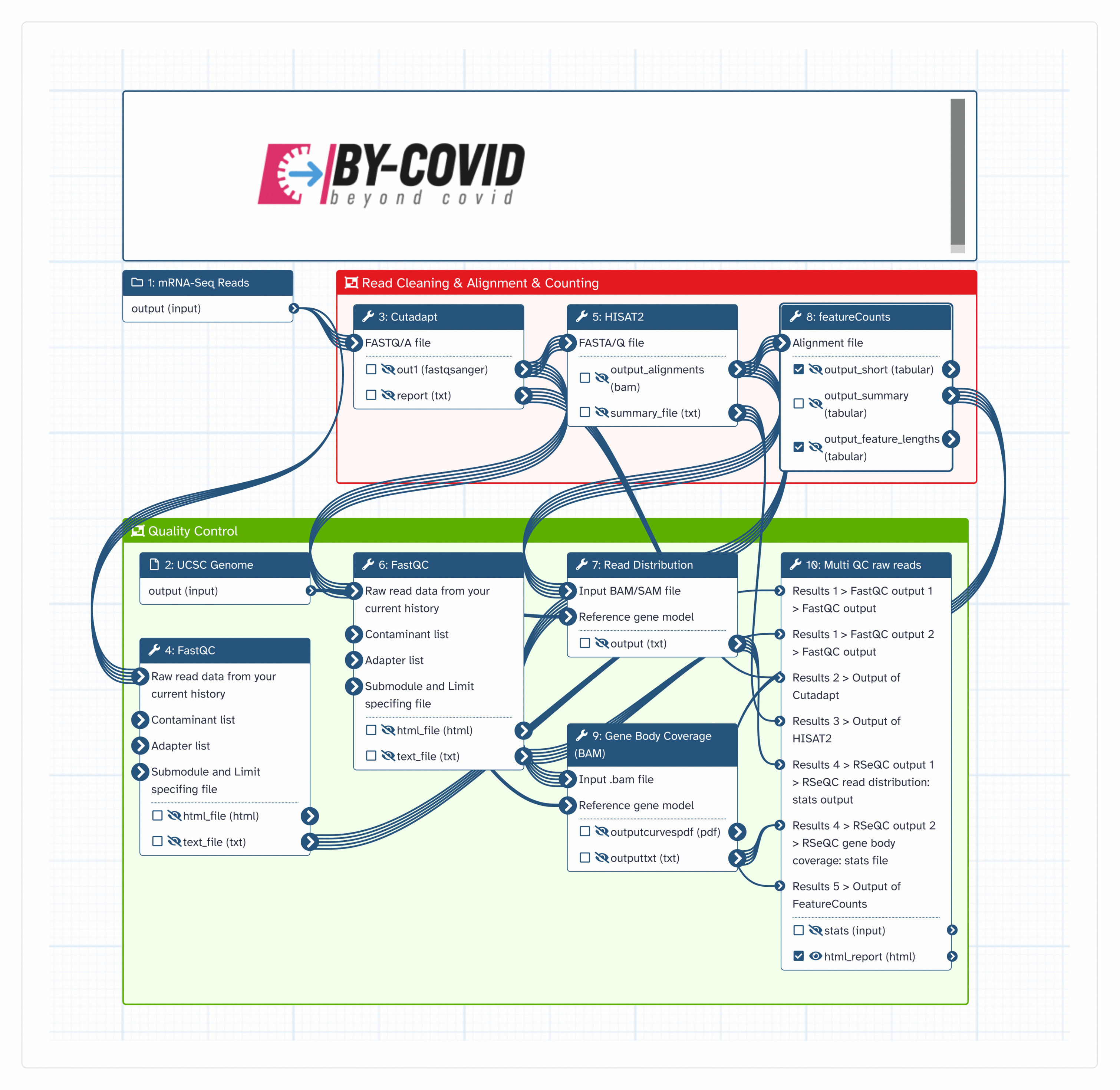
This portion of the workflow produces sets of feature Counts ready for analysis by limma/etc.
Inputs
| ID | Name | Description | Type |
|---|---|---|---|
| UCSC Genome | #main/UCSC Genome | Export of UCSC Genome, just the genes. |
|
| mRNA-Seq Reads | #main/mRNA-Seq Reads | Input list of fastqsanger format sequencing data |
|
Steps
| ID | Name | Description |
|---|---|---|
| 2 | FastQC | toolshed.g2.bx.psu.edu/repos/devteam/fastqc/fastqc/0.74+galaxy0 |
| 3 | Cutadapt | toolshed.g2.bx.psu.edu/repos/lparsons/cutadapt/cutadapt/4.4+galaxy0 |
| 4 | FastQC | toolshed.g2.bx.psu.edu/repos/devteam/fastqc/fastqc/0.74+galaxy0 |
| 5 | HISAT2 | toolshed.g2.bx.psu.edu/repos/iuc/hisat2/hisat2/2.2.1+galaxy1 |
| 6 | featureCounts | toolshed.g2.bx.psu.edu/repos/iuc/featurecounts/featurecounts/2.0.3+galaxy1 |
| 7 | Read Distribution | toolshed.g2.bx.psu.edu/repos/nilesh/rseqc/rseqc_read_distribution/5.0.1+galaxy2 |
| 8 | Gene Body Coverage (BAM) | toolshed.g2.bx.psu.edu/repos/nilesh/rseqc/rseqc_geneBody_coverage/5.0.1+galaxy2 |
| 9 | Multi QC raw reads | toolshed.g2.bx.psu.edu/repos/iuc/multiqc/multiqc/1.11+galaxy1 |
Outputs
| ID | Name | Description | Type |
|---|---|---|---|
| html_report | #main/html_report | n/a |
|
| output_feature_lengths | #main/output_feature_lengths | n/a |
|
| output_short | #main/output_short | n/a |
|
Version History
Version 1 (earliest) Created 19th Dec 2023 at 10:10 by Helena Rasche
Initial commit
Frozen
 Version-1
Version-17f98d5d
 Creators and Submitter
Creators and SubmitterCreators
Additional credit
Clinical Bioinformatics Unit, Pathology Department, Eramus Medical Center
Submitter
Activity
Views: 5339 Downloads: 1013 Runs: 1
Created: 19th Dec 2023 at 10:09
 Attributions
AttributionsNone
 Run on Galaxy
Run on Galaxy https://orcid.org/0000-0001-9760-8992
https://orcid.org/0000-0001-9760-8992 Download
Download