Workflow Type: Shell Script
Open
Work-in-progress
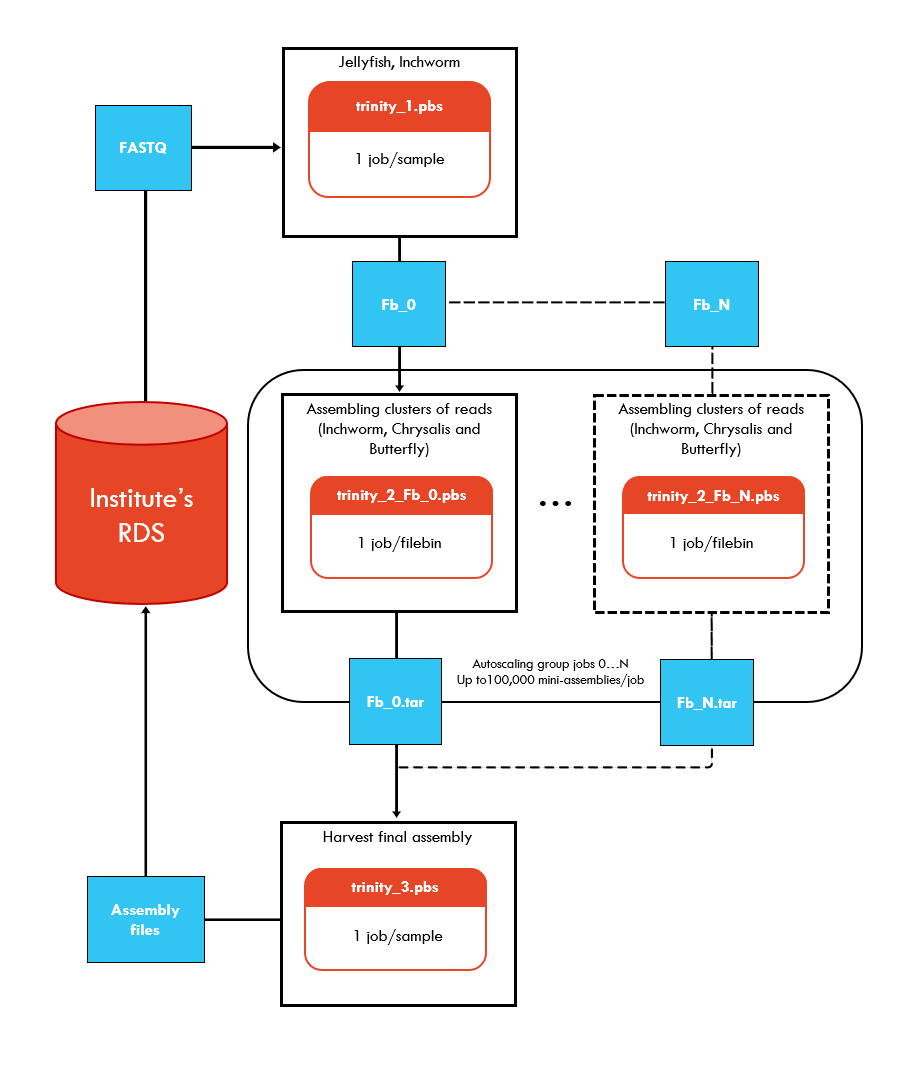
Description: Trinity @ NCI-Gadi contains a staged Trinity workflow that can be run on the National Computational Infrastructure’s (NCI) Gadi supercomputer. Trinity performs de novo transcriptome assembly of RNA-seq data by combining three independent software modules Inchworm, Chrysalis and Butterfly to process RNA-seq reads. The algorithm can detect isoforms, handle paired-end reads, multiple insert sizes and strandedness.
Infrastructure_deployment_metadata: Gadi (NCI)
Version History
Version 1 (earliest) Created 17th Aug 2021 at 05:44 by Tracy Chew
Added/updated 2 files
Open
 master
masteraf40e72
 Creators and Submitter
Creators and SubmitterCreators
Additional credit
Matthew Downton, Andrey Bliznyuk, Rika Kobayashi, Ben Menadue, Ben Evans
Submitter
Citation
Sadsad, R., Samaha, G., & Chew, T. (2021). Trinity @ NCI-Gadi. WorkflowHub. https://doi.org/10.48546/WORKFLOWHUB.WORKFLOW.145.1
License
Activity
Views: 4939 Downloads: 829
Created: 17th Aug 2021 at 05:44
Last updated: 7th Sep 2021 at 07:20
Annotated Properties
Topic annotations
Operation annotations
 Attributions
AttributionsNone
 Collections
Collections View on GitHub
View on GitHub https://orcid.org/0000-0003-2488-953X
https://orcid.org/0000-0003-2488-953X