Workflow Type: Galaxy
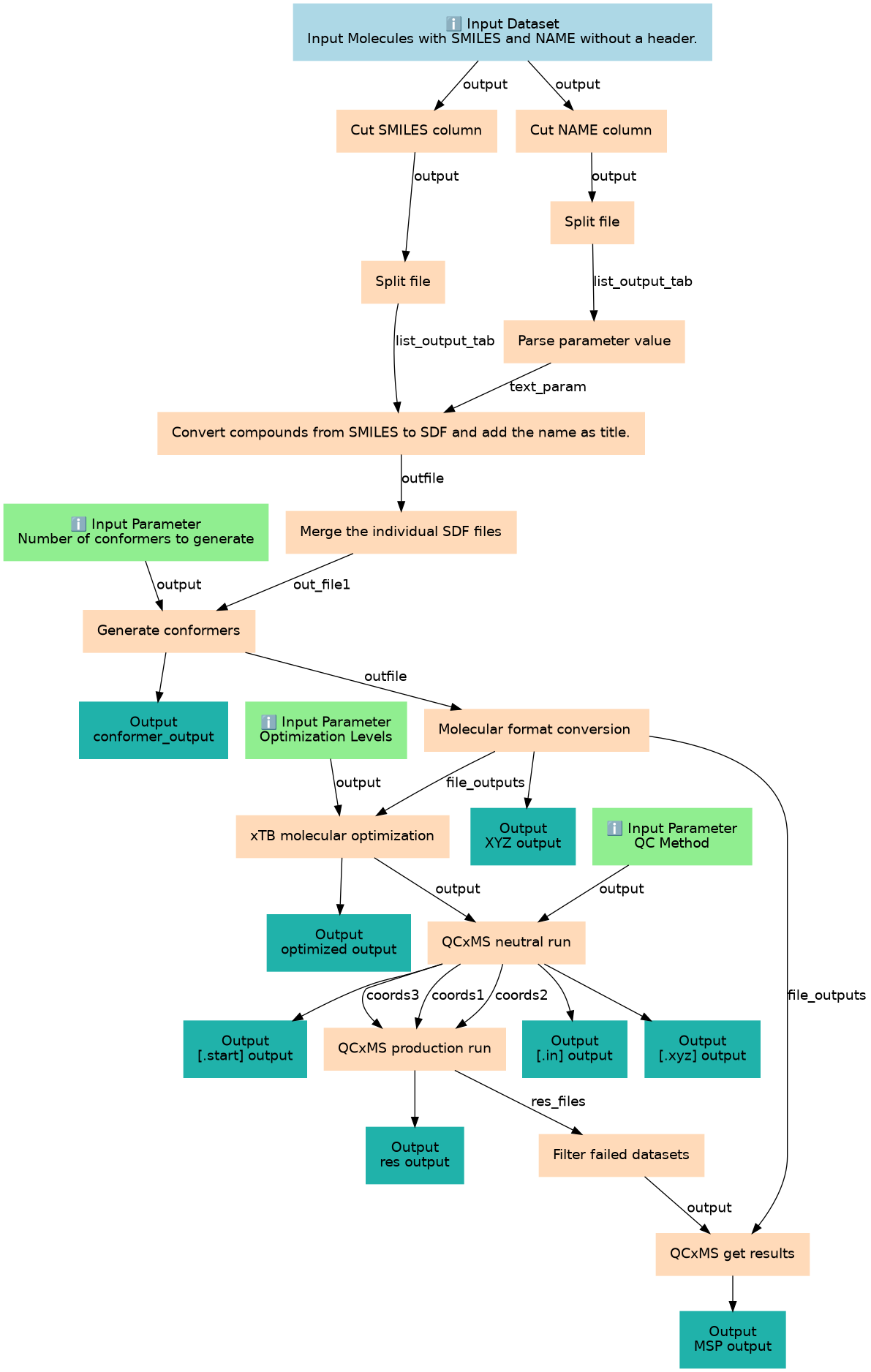
This workflow predict in-silico mass spectra using semi-empirical quantum physics method.
Associated Tutorial
This workflows is part of the tutorial Predicting EI+ mass spectra with QCxMS, available in the GTN
Features
- Includes Galaxy Workflow Tests
- Includes a Galaxy Workflow Report
- Uses Galaxy Workflow Comments
Thanks to...
Workflow Author(s): RECETOX SpecDat
Tutorial Author(s): Julia Jakiela, Helge Hecht
Tutorial Contributor(s): Saskia Hiltemann, Melanie Föll, Julia Jakiela
Inputs
| ID | Name | Description | Type |
|---|---|---|---|
| Input Molecules with SMILES and NAME without a header. | Input Molecules with SMILES and NAME without a header. | First column should containe the name of the molecule, the second should contain the SMILES code. |
|
| Number of conformers to generate | Number of conformers to generate | By default one conformer |
|
| Optimization Levels | Optimization Levels | Level of accuracy for the optimization |
|
| QC Method | QC Method | Available: GFN1-xTB and GFN2-xTB |
|
Steps
| ID | Name | Description |
|---|---|---|
| 4 | Cut SMILES column | toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_cut_tool/9.3+galaxy1 |
| 5 | Cut NAME column | toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_cut_tool/9.3+galaxy1 |
| 6 | Split file | toolshed.g2.bx.psu.edu/repos/bgruening/split_file_to_collection/split_file_to_collection/0.5.2 |
| 7 | Split file | toolshed.g2.bx.psu.edu/repos/bgruening/split_file_to_collection/split_file_to_collection/0.5.2 |
| 8 | Parse parameter value | param_value_from_file |
| 9 | Convert compounds from SMILES to SDF and add the name as title. | toolshed.g2.bx.psu.edu/repos/bgruening/openbabel_compound_convert/openbabel_compound_convert/3.1.1+galaxy0 |
| 10 | Merge the individual SDF files | toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_cat/9.3+galaxy1 |
| 11 | Generate conformers | Generate 3D conformers from SDF input for each molecule. It requires the number of conformers as an input parameter. Default parameters value is 1. toolshed.g2.bx.psu.edu/repos/bgruening/ctb_im_conformers/ctb_im_conformers/1.1.4+galaxy0 |
| 12 | Molecular format conversion | Convert the conformer to cartesian coordinate (XYZ) format toolshed.g2.bx.psu.edu/repos/bgruening/openbabel_compound_convert/openbabel_compound_convert/3.1.1+galaxy0 |
| 13 | xTB molecular optimization | Semi-empirical optimization toolshed.g2.bx.psu.edu/repos/recetox/xtb_molecular_optimization/xtb_molecular_optimization/6.6.1+galaxy3 |
| 14 | QCxMS neutral run | Produce preparation input files for production runs toolshed.g2.bx.psu.edu/repos/recetox/qcxms_neutral_run/qcxms_neutral_run/5.2.1+galaxy4 |
| 15 | QCxMS production run | Calculate the mass spectra for a given molecule using QCxMS. It generates .res files, which are collected and converted into MSP format in the last step toolshed.g2.bx.psu.edu/repos/recetox/qcxms_production_run/qcxms_production_run/5.2.1+galaxy3 |
| 16 | Filter failed datasets | Remove failed runs __FILTER_FAILED_DATASETS__ |
| 17 | QCxMS get results | Produce simulated mass spectra into MSP file format. toolshed.g2.bx.psu.edu/repos/recetox/qcxms_getres/qcxms_getres/5.2.1+galaxy2 |
Outputs
| ID | Name | Description | Type |
|---|---|---|---|
| conformer_output | conformer_output | n/a |
|
| XYZ output | XYZ output | n/a |
|
| optimized output | optimized output | n/a |
|
| [.in] output | [.in] output | n/a |
|
| [.xyz] output | [.xyz] output | n/a |
|
| [.start] output | [.start] output | n/a |
|
| res output | res output | n/a |
|
| MSP output | MSP output | n/a |
|
Version History
 Creators and Submitter
Creators and SubmitterCreators
Not specifiedSubmitter
Discussion Channel
Tool
License
Activity
Views: 1429 Downloads: 218 Runs: 0
Created: 2nd Jun 2025 at 11:00
 Attributions
AttributionsNone
 Visit source
Visit source Run on Galaxy
Run on Galaxy
 master
master