Workflow Type: Galaxy
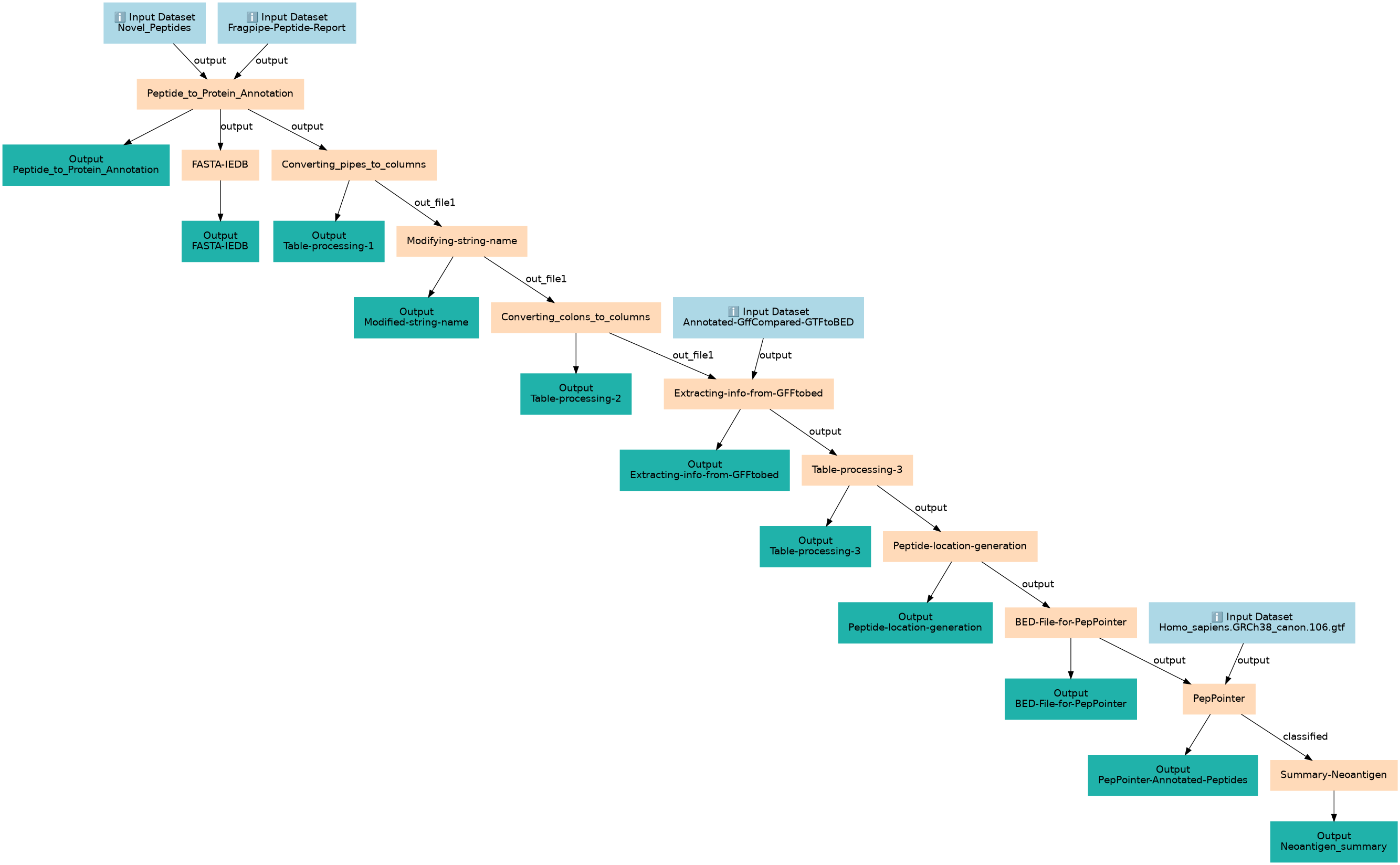
Annotating the novel peptides
Associated Tutorial
This workflows is part of the tutorial Neoantigen 4: Variant Annotation, available in the GTN
Features
- Includes a Galaxy Workflow Report
Thanks to...
Workflow Author(s): GalaxyP
Tutorial Author(s): Subina Mehta, Katherine Do, James Johnson
Tutorial Contributor(s): Pratik Jagtap, Timothy J. Griffin, Subina Mehta, Saskia Hiltemann, Katherine Do
Inputs
| ID | Name | Description | Type |
|---|---|---|---|
| Annotated-GffCompared-GTFtoBED | #main/Annotated-GffCompared-GTFtoBED | Annotated GffCompared GTF to BED |
|
| Fragpipe-Peptide-Report | #main/Fragpipe-Peptide-Report | Peptide report from Fragpipe workflow |
|
| Homo_sapiens.GRCh38_canon.106.gtf | #main/Homo_sapiens.GRCh38_canon.106.gtf | Homo_sapiens.GRCh38_canon.106.gtf |
|
| Novel_Peptides | #main/Novel_Peptides | Candidate Neoantigens from Fragpipe Workflow |
|
Steps
| ID | Name | Description |
|---|---|---|
| 4 | Peptide_to_Protein_Annotation | Extracting the information from Fragpipe for the Novel peptides toolshed.g2.bx.psu.edu/repos/iuc/query_tabular/query_tabular/3.3.2 |
| 5 | FASTA-IEDB | FASTA file for IEDB toolshed.g2.bx.psu.edu/repos/devteam/tabular_to_fasta/tab2fasta/1.1.1 |
| 6 | Converting_pipes_to_columns | converting pipes to columns Convert characters1 |
| 7 | Modifying-string-name | Modifying-string-name toolshed.g2.bx.psu.edu/repos/galaxyp/regex_find_replace/regexColumn1/1.0.3 |
| 8 | Converting_colons_to_columns | converting colons to columns Convert characters1 |
| 9 | Extracting-info-from-GFFtobed | Extracting-info-from-GFFtobed toolshed.g2.bx.psu.edu/repos/iuc/query_tabular/query_tabular/3.3.2 |
| 10 | Table-processing-3 | multiplying start and stop of the peptide location with 3 to calculate genomic coordinates toolshed.g2.bx.psu.edu/repos/iuc/query_tabular/query_tabular/3.3.2 |
| 11 | Peptide-location-generation | calculation genomic coordinates looking at peptide location toolshed.g2.bx.psu.edu/repos/iuc/query_tabular/query_tabular/3.3.2 |
| 12 | BED-File-for-PepPointer | Extracting bed file for Peppointer toolshed.g2.bx.psu.edu/repos/iuc/query_tabular/query_tabular/3.3.2 |
| 13 | PepPointer | PepPointer to add annotation to the peptides at Genomic level toolshed.g2.bx.psu.edu/repos/galaxyp/pep_pointer/pep_pointer/0.1.3+galaxy1 |
| 14 | Summary-Neoantigen | Summary file for Neoantigen toolshed.g2.bx.psu.edu/repos/iuc/query_tabular/query_tabular/3.3.2 |
Outputs
| ID | Name | Description | Type |
|---|---|---|---|
| BED-File-for-PepPointer | #main/BED-File-for-PepPointer | n/a |
|
| Extracting-info-from-GFFtobed | #main/Extracting-info-from-GFFtobed | n/a |
|
| FASTA-IEDB | #main/FASTA-IEDB | n/a |
|
| Modified-string-name | #main/Modified-string-name | n/a |
|
| Neoantigen_summary | #main/Neoantigen_summary | n/a |
|
| PepPointer-Annotated-Peptides | #main/PepPointer-Annotated-Peptides | n/a |
|
| Peptide-location-generation | #main/Peptide-location-generation | n/a |
|
| Peptide_to_Protein_Annotation | #main/Peptide_to_Protein_Annotation | n/a |
|
| Table-processing-1 | #main/Table-processing-1 | n/a |
|
| Table-processing-2 | #main/Table-processing-2 | n/a |
|
| Table-processing-3 | #main/Table-processing-3 | n/a |
|
Version History
 Creators and Submitter
Creators and SubmitterCreators
Not specifiedSubmitter
Discussion Channel
Activity
Views: 812 Downloads: 149 Runs: 0
Created: 7th Jul 2025 at 14:15
 Tags
Tags Attributions
AttributionsNone
 Visit source
Visit source Run on Galaxy
Run on Galaxy
 master
master