Workflow Type: Galaxy
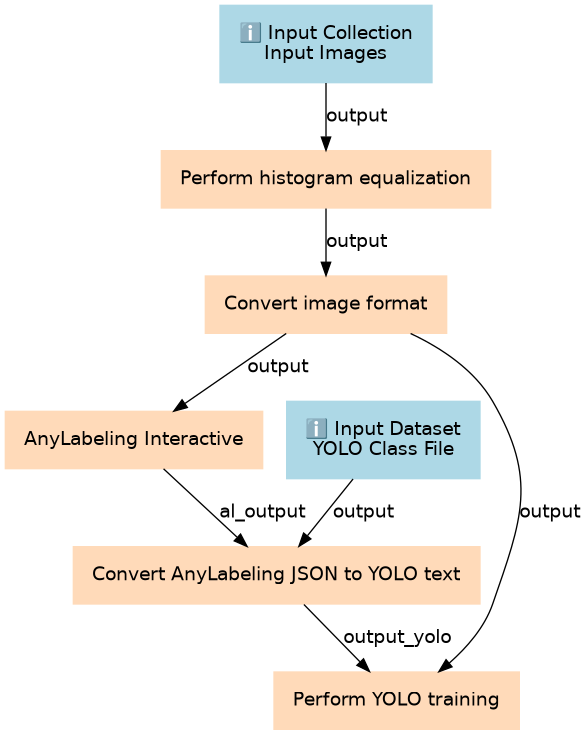
YOLO based tail analysis workflow for image segmentation and detection. The workflow is based on https://git.embl.org/grp-cba/tail-analysis/-/blob/main/analysis_workflow.md
Associated Tutorial
This workflows is part of the tutorial Training Custom YOLO Models for Object Detection and Segmentation in Bioimages, available in the GTN
Features
- Includes a Galaxy Workflow Report
Thanks to...
Workflow Author(s): nfdi4bioimage, Yi Sun, Arif Khan
Tutorial Author(s): Yi Sun, Arif UI Maula Khan
Inputs
| ID | Name | Description | Type |
|---|---|---|---|
| Input Images | Input Images | n/a |
|
| YOLO Class File | YOLO Class File | n/a |
|
Steps
| ID | Name | Description |
|---|---|---|
| 2 | Perform histogram equalization | toolshed.g2.bx.psu.edu/repos/imgteam/2d_histogram_equalization/ip_histogram_equalization/0.18.1+galaxy0 |
| 3 | Convert image format | toolshed.g2.bx.psu.edu/repos/bgruening/graphicsmagick_image_convert/graphicsmagick_image_convert/1.3.45+galaxy0 |
| 4 | AnyLabeling Interactive | interactive_tool_anylabeling |
| 5 | Convert AnyLabeling JSON to YOLO text | toolshed.g2.bx.psu.edu/repos/bgruening/json2yolosegment/json2yolosegment/8.3.0+galaxy2 |
| 6 | Perform YOLO training | toolshed.g2.bx.psu.edu/repos/bgruening/yolo_training/yolo_training/8.3.0+galaxy2 |
Version History
 Creators and Submitter
Creators and SubmitterCreators
Not specifiedSubmitter
Discussion Channel
Tool
Activity
Views: 970 Downloads: 147 Runs: 0
Created: 28th Jul 2025 at 14:17
 Attributions
AttributionsNone
 Visit source
Visit source Run on Galaxy
Run on Galaxy
 master
master