Workflow Type: Galaxy
Frozen
Frozen
Frozen
Stable
Galaxy Workflow Documentation: In-Silico Mass Spectra Prediction Using Semi-Empirical Quantum Chemistry
Overview
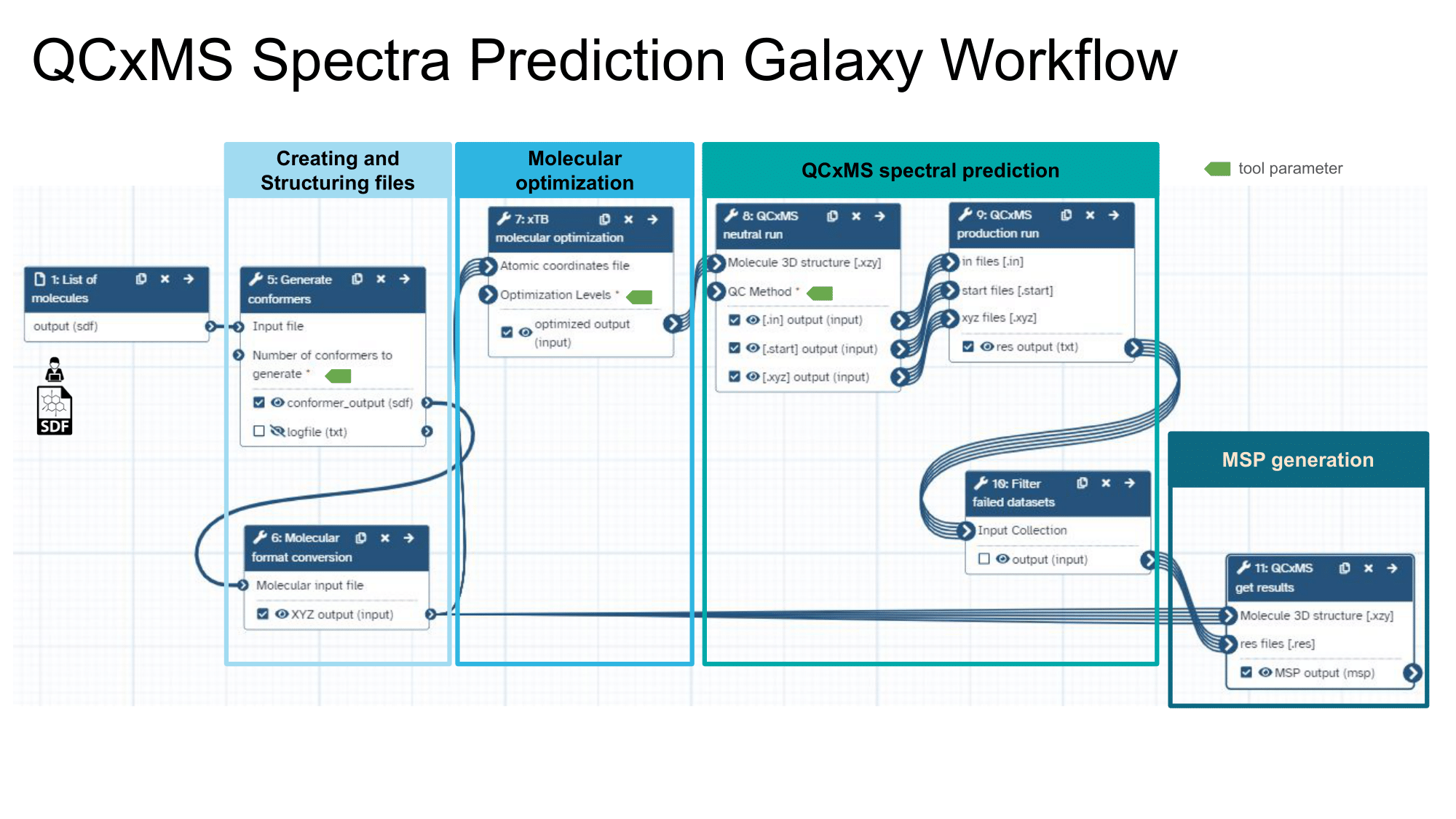
This workflow predicts in-silico mass spectra using a semi-empirical quantum chemistry method. It involves generating and optimizing molecular conformers and simulating their mass spectra with computational chemistry tools. The workflow receives an SDF file as input and outputs the mass spectrum in MSP file format.
Inputs
| ID | Name | Description | Type |
|---|---|---|---|
| List of molecules | #main/List of molecules | Global input dataset |
|
| Number of conformers to generate | #main/Number of conformers to generate | By default one conformer |
|
| Optimization Levels | #main/Optimization Levels | Level of accuracy for the optimization |
|
| QC Method | #main/QC Method | Available: GFN1-xTB and GFN2-xTB |
|
Steps
| ID | Name | Description |
|---|---|---|
| 4 | Generate conformers | Generate 3D conformers from SDF input for each molecule. It requires the number of conformers as an input parameter. Default parameters value is 1. toolshed.g2.bx.psu.edu/repos/bgruening/ctb_im_conformers/ctb_im_conformers/1.1.4+galaxy0 |
| 5 | Molecular format conversion | Convert the conformer to cartesian coordinate (XYZ) format toolshed.g2.bx.psu.edu/repos/bgruening/openbabel_compound_convert/openbabel_compound_convert/3.1.1+galaxy0 |
| 6 | xTB molecular optimization | Semi-empirical optimization toolshed.g2.bx.psu.edu/repos/recetox/xtb_molecular_optimization/xtb_molecular_optimization/6.6.1+galaxy1 |
| 7 | QCxMS neutral run | Produce preparation input files for production runs toolshed.g2.bx.psu.edu/repos/recetox/qcxms_neutral_run/qcxms_neutral_run/5.2.1+galaxy3 |
| 8 | QCxMS production run | Calculate the mass spectra for a given molecule using QCxMS. It generates .res files, which are collected and converted into MSP format in the last step toolshed.g2.bx.psu.edu/repos/recetox/qcxms_production_run/qcxms_production_run/5.2.1+galaxy2 |
| 9 | Filter failed datasets | Remove failed runs __FILTER_FAILED_DATASETS__ |
| 10 | QCxMS get results | Produce simulated mass spectra into MSP file format. toolshed.g2.bx.psu.edu/repos/recetox/qcxms_getres/qcxms_getres/5.2.1+galaxy1 |
Outputs
| ID | Name | Description | Type |
|---|---|---|---|
| MSP output | #main/MSP output | n/a |
|
| XYZ output | #main/XYZ output | n/a |
|
| [.in] output | #main/[.in] output | n/a |
|
| [.start] output | #main/[.start] output | n/a |
|
| [.xyz] output | #main/[.xyz] output | n/a |
|
| conformer_output | #main/conformer_output | n/a |
|
| optimized output | #main/optimized output | n/a |
|
| res output | #main/res output | n/a |
|
Version History
Version 3 (latest) Created 28th Apr 2025 at 09:47 by Helge Hecht
Updated QCxMS to fix a bug in parsing the electron temperature.
Frozen
 Version-3
Version-385c56dc
Galaxy Workflow End-to-end EI mass spectra prediction workflow using QCxMS Created 5th Aug 2024 at 14:53 by Helge Hecht
New version starting from a table with SMILES and NAMES to generate an SDF and then run the previous workflow.
Frozen
 Galaxy-Workflow-End-to-end-EI-mass-spectra-prediction-workflow-using-QCxMS
Galaxy-Workflow-End-to-end-EI-mass-spectra-prediction-workflow-using-QCxMS1a50bdb
Version 1 (earliest) Created 3rd Jun 2024 at 14:52 by Wudmir Rojas
qcxms galaxy workflow
Frozen
 Version-1
Version-10007a6a
 Creators and Submitter
Creators and SubmitterCreators
Additional credit
RECETOX SpecDat
Submitter
Tools
Citation
Ahmad, Z., Hecht, H., & Rojas, W. (2024). End-to-end spectra predictions: multi atoms dataset. WorkflowHub. https://doi.org/10.48546/WORKFLOWHUB.WORKFLOW.897.1
License
Activity
Views: 5458 Downloads: 1036 Runs: 15
Created: 3rd Jun 2024 at 14:52
Last updated: 28th Apr 2025 at 09:54
Annotated Properties
 Tags
Tags Attributions
AttributionsNone
 Run on Galaxy
Run on Galaxy https://orcid.org/0000-0001-6744-996X
https://orcid.org/0000-0001-6744-996X Download
Download