Workflow Type: Galaxy
Stable
Functional annotation of protein sequences
Associated Tutorial
This workflows is part of the tutorial Functional annotation of protein sequences, available in the GTN
Features
- Includes Galaxy Workflow Tests
Thanks to...
Workflow Author(s): Anthony Bretaudeau
Tutorial Author(s): Anthony Bretaudeau
Tutorial Contributor(s): Helena Rasche, Björn Grüning, Saskia Hiltemann, Bérénice Batut
Funder(s): ELIXIR Europe, Institut Français de Bioinformatique
Grants(s): Gallantries: Bridging Training Communities in Life Science, Environment and Health, EuroScienceGateway
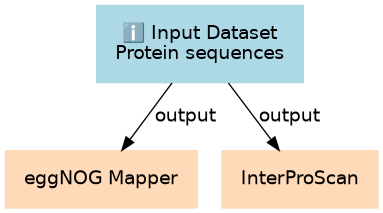
Inputs
| ID | Name | Description | Type |
|---|---|---|---|
| Protein sequences | #main/Protein sequences | Some protein sequences to functionally annotate |
|
Steps
| ID | Name | Description |
|---|---|---|
| 1 | eggNOG Mapper | toolshed.g2.bx.psu.edu/repos/galaxyp/eggnog_mapper/eggnog_mapper/2.1.8+galaxy3 |
| 2 | InterProScan | toolshed.g2.bx.psu.edu/repos/bgruening/interproscan/interproscan/5.59-91.0+galaxy3 |
Outputs
| ID | Name | Description | Type |
|---|---|---|---|
| _anonymous_output_1 | #main/_anonymous_output_1 | n/a |
|
| _anonymous_output_2 | #main/_anonymous_output_2 | n/a |
|
Version History
 Creators and Submitter
Creators and SubmitterCreator
Submitter
Discussion Channel
Citation
Bretaudeau, A. (2025). Functional annotation. WorkflowHub. https://doi.org/10.48546/WORKFLOWHUB.WORKFLOW.1521.1
Activity
Views: 1454 Downloads: 185 Runs: 0
Created: 2nd Jun 2025 at 10:57
Last updated: 28th Jul 2025 at 10:55
 Tags
Tags Attributions
AttributionsNone
 Collections
Collections Visit source
Visit source Run on Galaxy
Run on Galaxy
 1.0
1.0 https://orcid.org/0000-0003-0914-2470
https://orcid.org/0000-0003-0914-2470