Workflow Type: Common Workflow Language
Open
Work-in-progress
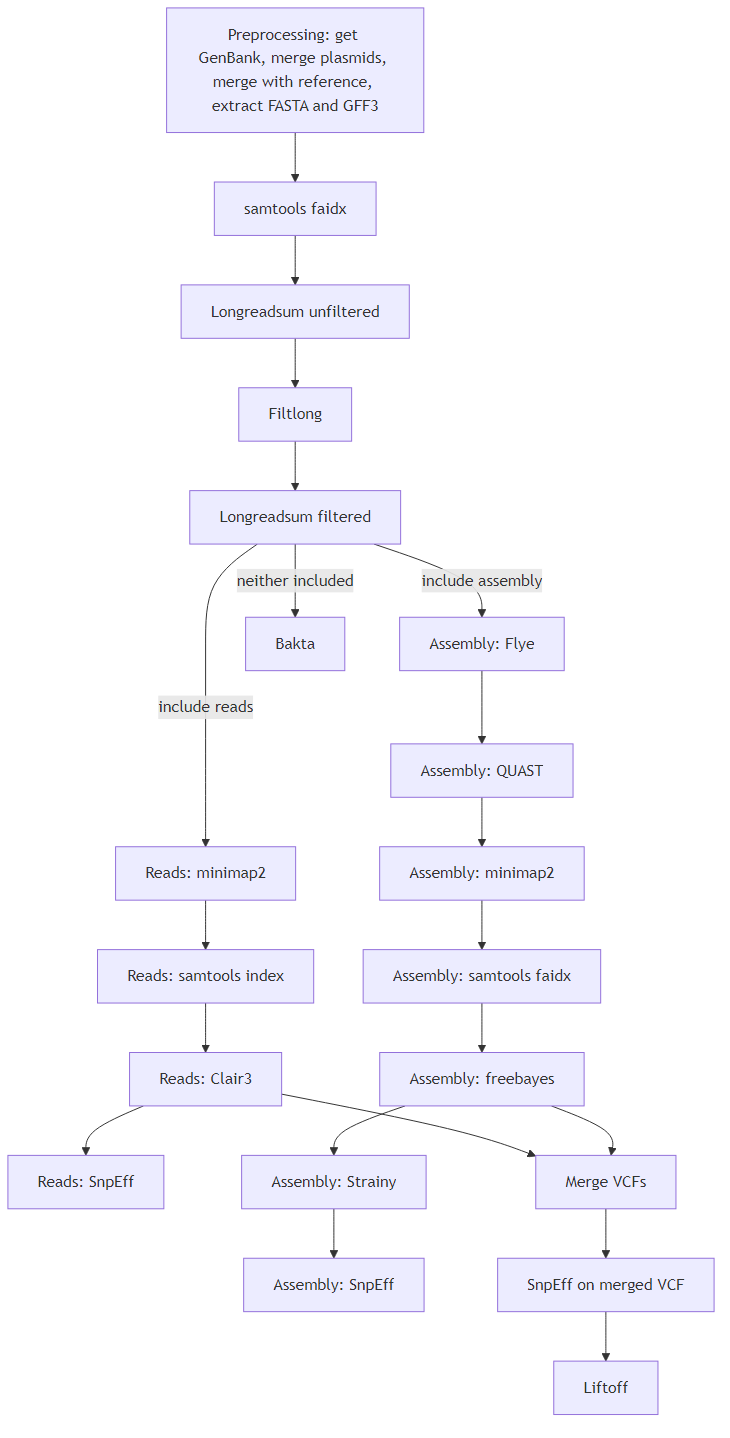
Workflow for long read quality control, contamination filtering, assembly, variant calling and annotation.
Steps:
- Preprocessing of reference file : https://workflowhub.eu/workflows/1818
- LongReadSum before and after filtering (read quality control)
- Filtlong filter on quality and length
- Flye assembly
- Minimap2 mapping of reads and assembly
- Clair3 variant calling of reads
- Freebayes variant calling of assembly
- Optional Bakta annotation of genomes with no reference
- SnpEff building or downloading of a database
- SnpEff functional annotation
- Liftoff annotation lift over
All tool CWL files and other workflows can be found here: Tools: https://git.wur.nl/ssb/automated-data-analysis/cwl/-/tree/main/tools Workflows: https://git.wur.nl/ssb/automated-data-analysis/cwl/-/tree/main/workflows
Inputs
| ID | Name | Description | Type |
|---|---|---|---|
| NCBI_identifier | NCBI genome identifier | NCBI Identifier of a genome for SnpEff to extract a genbank file and build a custom database out of. |
|
| annotation_file | annotation file to lift over | GFF or GTF file containing the annotations to lift over. |
|
| bakta_db | Bakta DB | Bakta database directory (default bakta-db_v5.1-light built in the container). |
|
| coverage_threshold | assembly coverage | Reduced coverage for the initial disjointig assembly. If set, Flye will downsample the reads to the specified coverage before assembly. Useful for high-coverage datasets to reduce memory usage. If not set, Flye will use all available reads. |
|
| dummy_annotation_file | n/a | n/a |
|
| dummy_database_folder | n/a | n/a |
|
| genome_size | Genome size | Estimated genome size (for example, 5m or 2.6g). |
|
| haploid_sensitive | haploid calling mode | Set to true to enable haploid calling mode, this is an experimental flag. |
|
| include_assembly | include assembly | Will include mapping and variant calling an assembly in the pipeline, default is true. |
|
| include_reads | include filtered reads | Will include mapping and variant calling filtered reads in the pipeline, default is true. |
|
| include_snpeff | include SnpEff | Will include functional interpretation of variants with SnpEff in the pipeline, default is true. |
|
| include_strainy | include strainy | Will include strain level analysis on the filtered reads, default is false. |
|
| input_read | long reads input | Long read sequence file in FASTQ format. |
|
| input_type | input file type | Acceptable input types: fa FASTA file input fq FASTQ file input f5 FAST5 file input f5s FAST5 file input with signal statistics output seqtxt sequencing_summary.txt input bam BAM file input rrms RRMS BAM file input Defaults to FQ file in this workflow. |
|
| keep_percent | Maximum read length threshold | Maximum read length threshold (default 90). |
|
| length_weight | Length weigth | Weight given to the length score (default 10). |
|
| log_level | level of logging | Logging level (1: DEBUG, 2: INFO, 3: WARNING, 4: ERROR, 5: CRITICAL), defaults to 2. |
|
| maximum_length | maximum length | Maximum read length threshold. |
|
| merging_script | merging script | Python script that merges input from both Clair3 and freebayes. Passed externally within the git structure to avoid having to host a new python docker. |
|
| min_alt_count | min_alt_count | Require at least this count of observations supporting an alternate allele. Defaults to 1 in this pipeline. |
|
| min_mean_q | minimum mean quality | Minimum mean quality threshold. |
|
| min_window_q | minimum window quality | Minimum window quality threshold. |
|
| minimum_length | Minimum read length | Minimum read length threshold (default 1000). |
|
| model_path | Clair3 Model Directory | Path to the Clair3 model inside the Docker container. |
|
| ncbi_data_exists | existing NCBI data | The used genome has an existing NCBI identifier, instead of annotating genes, the genbank file from NCBI will be used to build a database. |
|
| no_downstream | no downstream changes | Set to true to omit downstream changes. |
|
| no_phasing_for_fa | no phasing in full alignment | Set to true to skip whatshap phasing in full alignment, this is an experimental flag. |
|
| no_upstream | no upstream changes | Set to true to omit upstream changes. |
|
| plasmids | plasmid file(s) | Input plasmid GenBank files, which will be merged with the reference. |
|
| ploidy | ploidy settings | Settings of the ploidy, for haploid organisms, set to 1 (default). |
|
| provenance | include provenance information | Will include metadata on tool performance of LongReadSum, Filtlong, and Flye, default is true. |
|
| readtype | read type | Type of read i.e. PacBio or Nanopore. Used for naming output files. Defaults to Nanopore for this workflow, other read types are untested. |
|
| reference_gb | reference GenBank file | Reference file in GenBank format. If not provided requires NCBI identifier. |
|
| sample_name | sample name | Sample name, by default is extracted from the file input. Used as output names for LongReadSum, Filtlong, and minimap2. |
|
| seed | random seed | Sets the random seed for reproducability. Using the same seed number for random seed. Default is set to 1. |
|
| skip_qc_filtered | skip LongReadSum after filtering | Skip LongReadSum analyses of filter input data, default is false. |
|
| skip_qc_unfiltered | skip LongReadSum before filtering | Skip LongReadSum analyses of unfiltered input data, default is false. |
|
| snpeff_database_exists | existing SnpEff database | The used genome has an existing database within SnpEff, instead of building a database, the existing database will be downloaded, default is false. |
|
| snpeff_genome | genome/database identifier | Identifier for the SnpEff database to download or build (e.g. 'GRCh37.75' for human, or a custom name for microbial strains). |
|
| target_bases | target bases | Keep only the best reads up to this many total bases. |
|
| threads | Number of threads | Number of threads to use for computational processes. |
|
| transfer_annotation | transfer annotation | Whether the annotation of the reference should be carried over to the new assembly (use Liftoff), default is false. |
|
Steps
| ID | Name | Description |
|---|---|---|
| bakta | bakta genome annotation | Bacterial genome annotation, only runs when no reference (genbank file(s) or NCBI identifier) is supplied. |
| clair3 | Clair3 variant calling | Variant calling of filtered reads with Clair3 using input models. |
| filtlong | long read filtering | Filter long reads based on set parameters. |
| filtlong_files_to_folder | Filtlong folder | Preparation of Filtlong output files to a specific output folder. |
| flye | Flye assembly | De novo assembly of single-molecule reads with Flye. |
| flye_files_to_folder | Flye output folder | Preparation of Flye output files to a specific output folder. |
| freebayes | FreeBayes variant calling | Variant calling of assembly with FreeBayes. |
| liftoff | Liftoff annotation lift over | Lifting over annotations from reference to assembly. |
| liftoff_files_to_folder | liftoff assembly output folder | Preparation of Liftoff output files to a specific output folder. |
| longreadsum_filtered | LongReadSum filtered | LongReadSum Quality assessment of reads after filtering. |
| longreadsum_unfiltered | LongReadSum unfiltered | LongReadSum Quality assessment of reads prior to filtering. |
| merging_vcfs | merging vcf files | Merging the VCF output from Clair3 and freebayes. |
| minimap2_assembly | Minimap2 assembly mapping | Assembly mapping of filtered reads using Minimap2. |
| minimap2_reads | Minimap2 read mapping | Read mapping of filtered reads using Minimap2. |
| preprocess_reference | plasmid preprocessing | Pre-processing of reference, merging reference with optional plasmid input and extracting GenBank, GFF3 and FASTA files. |
| provenance_files_to_folder | provenance output folder | Preparation of provenance output files to a specific output folder. |
| quast | QUAST quality assessment | Quality assessment of assembly with QUAST. |
| samtools_assembly_index | samtools index assembly | Indexing of assembly BAM file with samtools index. |
| samtools_faidx_assembly | samtools faidx assembly | Indexing of FASTA file with samtools faidx. |
| samtools_faidx_reads | samtools faidx | Indexing of FASTA file with samtools faidx. |
| samtools_reads_index | samtools index reads | Indexing of reads BAM file with samtools index. |
| snpeff_assembly | SnpEff assembly | Running SnpEff on the assembly variant output of freebayes. |
| snpeff_assembly_files_to_folder | SnpEff assembly output folder | Preparation of SnpEff assembly output files to a specific output folder. |
| snpeff_build | SnpEff database building | Downloading of a SnpEff database based on the genome name within the database. |
| snpeff_download | SnpEff database downloading | Downloading of a SnpEff database based on the genome name within the database. |
| snpeff_merged | SnpEff merged | Running SnpEff on the merged variant output of both Clair3 and freebayes. |
| snpeff_merged_files_to_folder | SnpEff merged output folder | Preparation of SnpEff merged output files to a specific output folder. |
| snpeff_reads | SnpEff reads | Running SnpEff on the reads variant output of Clair3. |
| snpeff_reads_files_to_folder | SnpEff reads output folder | Preparation of SnpEff reads output files to a specific output folder. |
| strainy | Strainy strain level analysis | Strain level analysis on assembled reads. Produces multi-allelic phasing, individual haplotypes and strain-specific variant calls. |
| unzip | unzipping clair3 | Unzipping Clair3 VCF file. |
Outputs
| ID | Name | Description | Type |
|---|---|---|---|
| assembly__fasta_index_out | indexed reference | Indexed reference FASTA file. |
|
| assembly_bam_index_out | indexed mapped assembly | Indexed mapped assembly. |
|
| bakta_outdir | bakta folder | Folder with bakta output files. |
|
| clair3_outdir | Clair3 output directory | Clair3 output directory containing the vcf file. |
|
| clair3_vcf | Clair3 output file | Output variant file from Clair3. |
|
| filtlong_outdir | Filtlong folder | Folder with Filtlong output files. |
|
| flye_outdir | Filtlong folder | Folder with Filtlong output files. |
|
| freebayes_output | freebayes output file | Output variant file from freebayes. |
|
| liftoff_outdir | Liftoff folder | Folder with liftoff output files. |
|
| logs_outdir | logs folder | Folder with provenance information. |
|
| longreadsum_filtered_outdir | LongReadSum folder 2 | Folder with LongReadSum output files. |
|
| longreadsum_unfiltered_outdir | LongReadSum folder | Folder with LongReadSum output files. |
|
| merged_output | merged output file | Merged output variant file from both Clair3 and freebayes. |
|
| minimap2_assembly_bam | mapped assembly | Assembly mapped by minimap2. |
|
| minimap2_reads_bam | mapped reads | Filtered reads mapped by minimap2. |
|
| preprocessed_fasta | preprocessed FASTA file | The preprocessed FASTA file. This file is extracted from the above GenBank file. |
|
| preprocessed_genbank | preprocessed GenBank file | The preprocessed GenBank file. This file only differs from the input GenBank file (if provided) when plasmids are included. |
|
| preprocessed_gff3 | preprocessed GFF3 file | The preprocessed GFF3 file. This file is extracted from the above GenBank file. |
|
| quast_outdir | Filtlong folder | Folder with Filtlong output files. |
|
| reads_bam_index_out | indexed mapped reads | Indexed filtered mapped reads. |
|
| reads_fasta_index_out | indexed reference | Indexed reference FASTA file. |
|
| snpeff_assembly_outdir | SnpEff assembly folder | Folder with SnpEff assembly output files. |
|
| snpeff_merged_outdir | SnpEff merged folder | Folder with SnpEff merged output files. |
|
| snpeff_reads_outdir | SnpEff reads folder | Folder with SnpEff reads output files. |
|
| strainy_outdir | strainy folder | Folder with strainy output files. |
|
Version History
Version 1 (earliest) Created 12th Aug 2025 at 13:00 by Martijn Melissen
Initial commit
Open
 master
master25fa72f
 Creators and Submitter
Creators and SubmitterCreator
Submitter
Activity
Views: 705 Downloads: 166
Created: 12th Aug 2025 at 13:00
Annotated Properties
Topic annotations
Operation annotations
 Attributions
AttributionsNone
 Visit source
Visit source https://orcid.org/0009-0005-0017-0928
https://orcid.org/0009-0005-0017-0928