Workflows for Galaxy Climate
Space: Independent Teams
SEEK ID: https://workflowhub.eu/projects/18
Public web page: Not specified
Organisms: No Organisms specified
WorkflowHub PALs: No PALs for this Team
Team created: 23rd Jul 2020
Related items
Teams: Galaxy Climate, Euro-BioImaging
Organizations: gx-climate, Euro-BioImaging
Teams: Galaxy Climate
Organizations: Galaxy
A space managed by WorkflowHub administrators for teams that don't want/need to manage their own space.
Teams: IBISBA Workflows, NMR Workflow, UNLOCK, NanoGalaxy, Galaxy Climate, PNDB, IMBforge, COVID-19 PubSeq: Public SARS-CoV-2 Sequence Resource, LBI-RUD, Nick-test-team, Italy-Covid-data-Portal, UX trial team, Integrated and Urban Plant Pathology Laboratory, SARS-CoV-2 Data Hubs, lmjxteam2, virAnnot pipeline, Ay Lab, iPC: individualizedPaediatricCure, Harkany Lab, MOLGENIS, EJPRD WP13 case-studies workflows, Common Workflow Language (CWL) community, Testing, SeBiMER, IAA-CSIC, MAB - ATGC, Probabilistic graphical models, GenX, Snakemake-Workflows, ODA, IPK BIT, CO2MICS Lab, FAME, CHU Limoges - UF9481 Bioinformatique / CNR Herpesvirus, Quadram Institute Bioscience - Bioinformatics, HecatombDevelopment, Institute of Human Genetics, Testing RO Crates, Test Team, Applied Computational Biology at IEG/HMGU, INFRAFRONTIER workflows, OME, TransBioNet, OpenEBench, Bioinformatics and Biostatistics (BIO2 ) Core, VIB Bioinformatics Core, CRC Cohort, ICAN, MustafaVoh, Single Cell Unit, CO-Graph, emo-bon, TestEMBL-EBIOntology, CINECA, Toxicology community, Pitagora-Network, Workflows Australia, Medizinisches Proteom-Center, Medical Bioinformatics, AGRF BIO, EU-Openscreen, X-omics, ELIXIR Belgium, URGI, Size Inc, GA-VirReport Team, The Boucher Lab, Air Quality Prediction, pyiron, CAPSID, Edinburgh Genomics, Defragmentation TS, NBIS, Phytoplankton Analysis, Seq4AMR, Workflow registry test, Read2Map, SKM3, ParslRNA-Seq: an efficient and scalable RNAseq analysis workflow for studies of differentiated gene expression, de.NBI Cloud, Meta-NanoSim, ILVO Plant Health, EMERGEN-BIOINFO, KircherLab, Apis-wings, BCCM_ULC, Dessimoz Lab, TRON gGmbH, GEMS at MLZ, Computational Science at HZDR, Big data in biomedicine, TRE-FX, MISTIC, Guigó lab, Statistical genetics, Delineating Regions-of-interest for Mass Spectrometry Imaging by Multimodally Corroborated Spatial Segmentation, OLCF-WES, Bioinformatics Unit @ CRG, Bioinformatics Innovation Lab, BSC-CES, ELIXIR Proteomics, Black Ochre Data Labs, Zavolan Lab, Metabolomics-Reproducibility, Team Cardio, NGFF Tools, Bioinformatics workflows for life science, Workflows for geographic science, Pacific-deep-sea-sponges-microbiome, CSFG, SNAKE, Katdetectr, INFRAFRONTIER GmbH, PerMedCoE, Euro-BioImaging, EOSC-Life WP3 OC Team, cross RI project, ANSES-Ploufragan, SANBI Pathogen Bioinformatics, Biodata Analysis Group, DeSci Labs, Erasmus MC - Viroscience Bioinformatics, ARA-dev, Mendel Centre for Plant Genomics and Proteomics, Metagenomic tools, WorkflowEng, Polygenic Score Catalog, bpm, scNTImpute, Systems Biotechnology laboratory, Cimorgh IT solutions, MLme: Machine Learning Made Easy, Hurwitz Lab, Dioscuri TDA, Scipion CNB, System Biotechnology laboratory, yPublish - Bioinfo tools, NIH CFDE Playbook Workflow Partnership, MMV-Lab, EMBL-Bioimage Analysis Support, EBP-Nor, Evaluation of Swin Transformer and knowledge transfer for denoising of super-resolution structured illumination microscopy data, Bioinformatics Laboratory for Genomics and Biodiversity (LBGB), multi-analysis dFC, CholGen, RNA group, Plant Genomes Pipelines in Galaxy, Pathogen Genomic Laboratory, Chemical Data Lab, JiangLab, Pangenome database project, HP2NET - Framework for construction of phylogenetic networks on High Performance Computing (HPC) environment, Center for Open Bioimage Analysis, Generalized Open-Source Workflows for Atomistic Molecular Dynamics Simulations of Viral Helicases, Historical DNA genome skimming, QCDIS, Peter Menzel's Team, NHM Clark group, ESRF Workflow System (Ewoks), Kalbe Bioinformatics, Nextflow4Metabolomics, GBCS, CEMCOF, Jackson Laboratory NGS-Ops, Schwartz Lab, BRAIN - Biomedical Research on Adult Intracranial Neoplasms, Cancer Therapeutics and Drug Safety, Deepdefense, Mid-Ohio Regional Planning Commission, MGSSB, Institute for Human Genetics and Genomic Medicine Aachen, FengTaoSMU, EGA, Plant-Food-Research-Open, KrauthammerLab, Geo Workflows, grassland pDT, FunGIALab, CRIM - Computer Research Institute of Montréal, Medvedeva Lab, Metagenlab, FAIR-EASE, Protein-protein and protein-nucleic acid binding site prediction research, Culhane Lab, IDUN - Drug Delivery and Sensing, Edge Computing DAG Task Scheduling Research Group, Stratum corneum nanotexture feature detection using deep learning and spatial analysis: a non-invasive tool for skin barrier assessment, COPO, Taudière group, ErasmusMC Clinical Bioinformatics, interTwin, fluid flow modeling, EnrichDO, WorkflowResearch, Application Security - Test Crypt4GH solutions, RenLabBioinformatics, Yongxin's team, PiFlow, HLee_SeoGroup, UFZ - Image Data Management and Processing Workflows, Korean Bioinformaticians, Into the deep, XChem, CPM, SocialGene, Research Data Management ICE-2, ObjectRecognition, LiDAR, FONDA II C2, Astroparticle Lab, FAIRagro M4.4, Kgerring, QuackenbushLab, Virus sequencing team, SOS, BioImage Informatics and Analysis Workflows, BoostNano, simblockflow, CSSB, Research on Workflow scheduling, Research Data1, CSUbioinformatics, CDPP, Mr., ASD-HRS, data management, FAIR_thesis: Marine acoustic data, CellBinDB, DEEP Lab, University of Amsterdam, SIMEXP, nf-pediatric Team, Kasmanas, Structural Variation Analysis, fuzzyworkflow, CausalCoxMGM Team, Tufts University Center for Cellular Agriculture (TUCCA), Test, CrustyBase, Applied Computational Cancer Research, Click-qPCR, BAID Team, FabianLab, Vector informatics and genomics group, AlmondBreedingLab, Artificial Design for Intelligent Breeding, ELIXIR Biodiversity Community, GROTIA, Biomedical_LLM, WhiteSymmetry, Hämatologie Labor Kiel, pakbaba, Metagenomics Analysis, MTB Bioinformatics Workflows, RTC Bioinformatics, CMG-GUTS, EI Papatheodorou Group, Metagenomics Analysis of Microbiome, ColoPola: A polarimetric imaging dataset for colorectal cancer detection, AI in Zhou Lab, CEPLAS, LTER-Italy, Gevaert Lab, Kendall-Theisen lab pipelines, High Performance Scientific Computing Laboratory (HPSC), Ensembl Metazoa and Plants, BiRD, Q-Dawn, AMRMALDI, Systems and Synthetic Biology, EI Core Bioinformatics Group, Values Shape Community, SolutionMake, GENEX, iPol, zxfenglab, Bioinformatics Unit IIS-FJD, omnibenchmark, BioX Fanatics, Li-Omics Lab, Metro Team, ZiemertLab, Song Lab, PanGIA, Holobionts Workflows, An Ideal Design for RNA-seq investigations into RPL, AMI2B Team, Institute for Hearing Technology and Acoustics (IHTA), Transcriptomics in unexplained recurrent pregnancy loss, E-MOBI / EKONOMIK MOBIL,S.R.L, Workflow Run Crate working group, datafun, BioFAIR, BH2025_Project30, NII Test Team, LIB, Galaxy and Beyond
Web page: Not specified
ROR ID: Not specified
Department: Not specified
Country:  Norway
Norway
City: Not specified
Web page: Not specified
Annual percentage Change for population in Germany (1950 - 2025)
This workflow run was executed on Galaxy (Workflow Rerun Information)
Workflow: Climate Stripes
Execution Status: scheduled
Executed: 2025-05-27 14:14:07.240149
Workflow Inputs
Formal Input Definitions
-
Germany-Population-Annual--Change-2025-05-27-15-17.csv (File)
-
Column name to use for plotting (Text)
-
Plot Title (Text)
-
nxsplit (Integer)
-
Description: Number of values per intervals
...
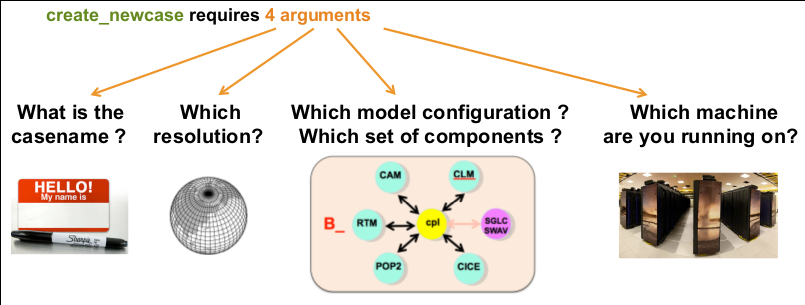
This workflow demonstrates the usage of the Community Earth System Model on Galaxy Europe.
A fully coupled B1850 compset with resolution f19_g17 is run for 1 month.
This workflow demonstrates the usage of EODIE, a toolkit to extract object based timeseries information from Earth Observation data.
EODIE is a toolkit to extract object based timeseries information from Earth Observation data.
The EODIE code can be found on Gitlab .
The goal of EODIE is to ease the extraction of time series information at object level. Today, vast amounts of Earth Observation data are available to the users via for example earth explorer ...
Abstract CWL Automatically generated from the Galaxy workflow file: GTN 'Pangeo 101 for everyone - Introduction to Xarray'.
In this tutorial, we analyze particle matter < 2.5 μm/m3 data from Copernicus Atmosphere Monitoring Service to understand Xarray Galaxy Tools:
- Understand how an Xarray dataset is organized;
- Get metadata from Xarray dataset such as variable names, units, coordinates (latitude, longitude, level), etc;
- Plot an Xarray dataset on a geographical map and learn to customize ...
This workflow extracts 5 different time periods e.g. January- June 2019, 2020 and 2021, July-December 2019 and 2020 over a single selected location. Then statistics (mean, minimum, maximum) are computed. The final products are maximum, minimum and mean.
This workflow is used to process timeseries from meteorological stations in Finland but can be applied to any timeseries according it follows the same format.
Take a temperature timeseries from any meteorological station. Input format is csv and it must be standardized with 6 columns:
- Year (ex: 2021)
- month (ex: 1)
- day (ex: 15)
- Time (ex: 16:56)
- Time zone (such as UTC)
- Air temperature (degC)
Abstract CWL Automatically generated from the Galaxy workflow file: CLM-FATES_ ALP1 simulation (5 years)
Description: SSP-based RCP scenario with high radiative forcing by the end of century. Following approximately RCP8.5 global forcing pathway with SSP5 socioeconomic conditions. Concentration-driven. Rationale: the scenario represents the high end of plausible future pathways. SSP5 is the only SSP with emissions high enough to produce the 8.5 W/m2 level of forcing in 2100.
This workflow is answering to the following scientific question:
- Is it worth investing in artificial snowmaking equipment ...
Abstract CWL Automatically generated from the Galaxy workflow file: Workflow with Copernicus Essential Climate Variable - select and plot
 Overview
Overview Tags
Tags Download
Download