Workflow Type: Galaxy
Open
Stable
Using:
- vadr annotation (virus model must be selected in options)
- vardict variant caller
- coverage depth Provides summarizing files:
- png image of variant calling with annotations and coverage depths
- tsv file with all information of significant variants only
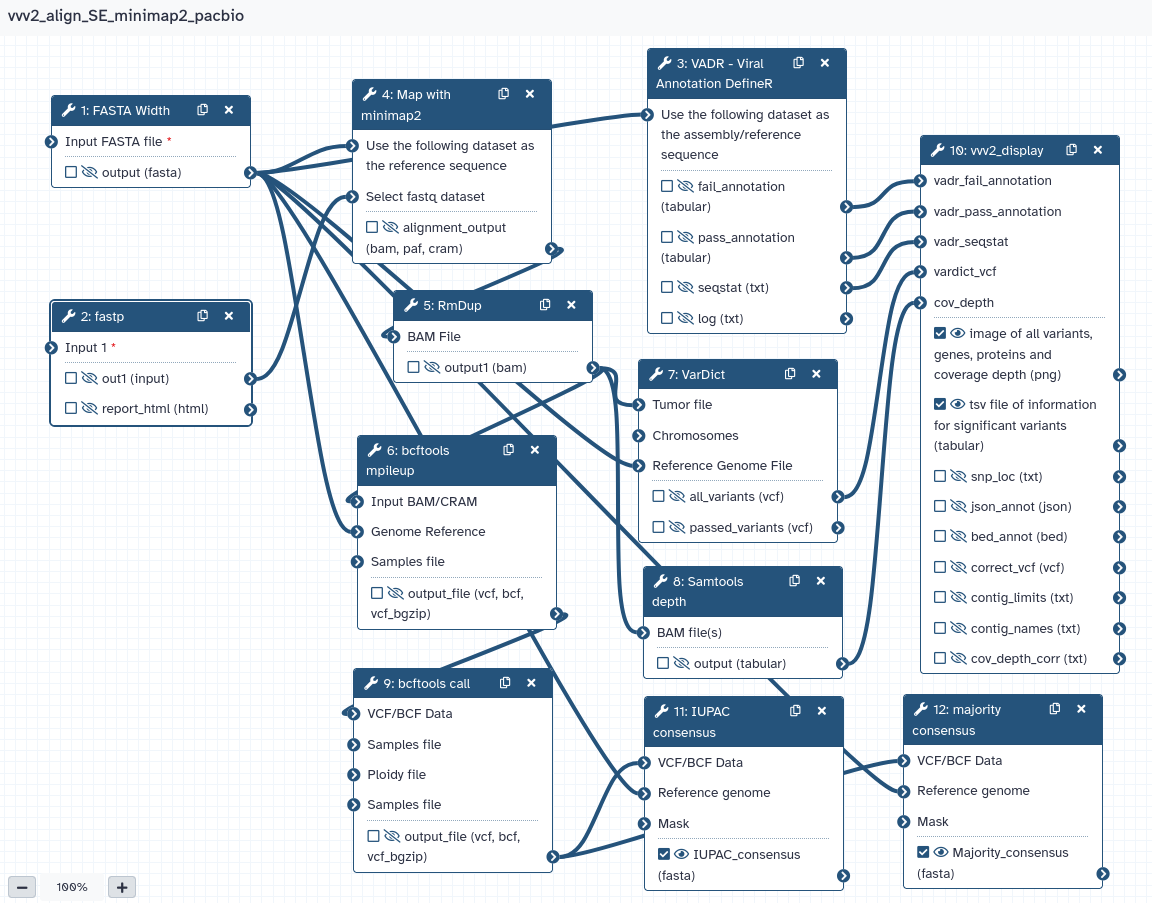
Steps
| ID | Name | Description |
|---|---|---|
| 0 | FASTA Width | toolshed.g2.bx.psu.edu/repos/devteam/fasta_formatter/cshl_fasta_formatter/1.0.1+galaxy2 |
| 1 | fastp | toolshed.g2.bx.psu.edu/repos/iuc/fastp/fastp/0.23.2+galaxy0 |
| 2 | VADR - Viral Annotation DefineR | toolshed.g2.bx.psu.edu/repos/ftouzain/vadr/vadr/0.2.0 |
| 3 | Map with minimap2 | toolshed.g2.bx.psu.edu/repos/iuc/minimap2/minimap2/2.28+galaxy1 |
| 4 | RmDup | toolshed.g2.bx.psu.edu/repos/devteam/samtools_rmdup/samtools_rmdup/2.0.1 |
| 5 | bcftools mpileup | toolshed.g2.bx.psu.edu/repos/iuc/bcftools_mpileup/bcftools_mpileup/1.15.1+galaxy3 |
| 6 | VarDict | toolshed.g2.bx.psu.edu/repos/iuc/vardict_java/vardict_java/1.8.3+galaxy1 |
| 7 | Samtools depth | toolshed.g2.bx.psu.edu/repos/iuc/samtools_depth/samtools_depth/1.15.1+galaxy2 |
| 8 | bcftools call | toolshed.g2.bx.psu.edu/repos/iuc/bcftools_call/bcftools_call/1.15.1+galaxy3 |
| 9 | vvv2_display | - png image of variants - tsv file of variants info toolshed.g2.bx.psu.edu/repos/ftouzain/vvv2_display/vvv2_display/0.2.4.0 |
| 10 | IUPAC consensus | toolshed.g2.bx.psu.edu/repos/iuc/bcftools_consensus/bcftools_consensus/1.15.1+galaxy3 |
| 11 | majority consensus | Gives consensus with major nucleotides only toolshed.g2.bx.psu.edu/repos/iuc/bcftools_consensus/bcftools_consensus/1.15.1+galaxy3 |
Outputs
| ID | Name | Description | Type |
|---|---|---|---|
| IUPAC_consensus | #main/IUPAC_consensus | n/a |
|
| Majority_consensus | #main/Majority_consensus | n/a |
|
| image of all variants, genes, proteins and coverage depth | #main/image of all variants, genes, proteins and coverage depth | n/a |
|
| tsv file of information for significant variants | #main/tsv file of information for significant variants | n/a |
|
Version History
Version 1 (earliest) Created 19th Jun 2025 at 11:27 by Fabrice Touzain
Initial commit
Open
 master
master88a9377
 Creators and Submitter
Creators and SubmitterCreator
Additional credit
This study was founded by the French National Research Agency and by Santé publique France as part of the project "EMERGEN". Anses Ploufragan research was also supported by Agglomération de Saint-Brieuc, Département des Côtes d'Armor and Région Bretagne
Submitter
Activity
Views: 1307 Downloads: 184 Runs: 3
Created: 19th Jun 2025 at 11:27
Last updated: 19th Jun 2025 at 11:29
 Attributions
AttributionsNone
 Run on Galaxy
Run on Galaxy https://orcid.org/0000-0002-1103-2425
https://orcid.org/0000-0002-1103-2425