Workflow Type: Galaxy
Open
Deprecated
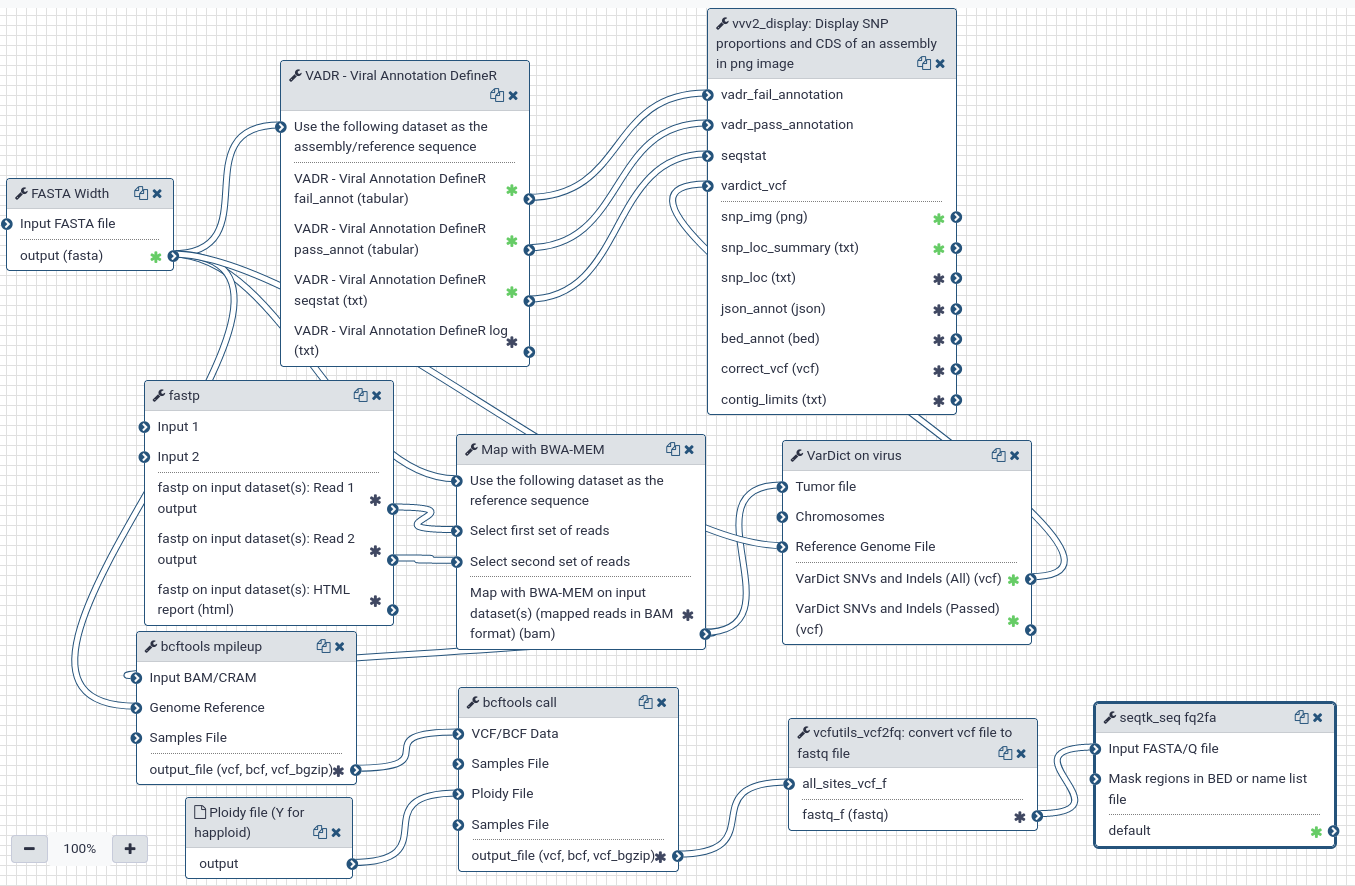
PAIRED-END workflow. Align reads on fasta reference/assembly using bwa mem, get a consensus, variants, mutation explanations.
IMPORTANT:
- For "bcftools call" consensus step, the --ploidy file is in "Données partagées" (Shared Data) and must be imported in your history to use the worflow by providing this file (tells bcftools to consider haploid variant calling).
- SELECT THE MOST ADAPTED VADR MODEL for annotation (see vadr parameters).
Inputs
| ID | Name | Description | Type |
|---|---|---|---|
| Ploidy file (Y for happloid) | Ploidy file (Y for happloid) | n/a |
|
Steps
| ID | Name | Description |
|---|---|---|
| 0 | FASTA Width | toolshed.g2.bx.psu.edu/repos/devteam/fasta_formatter/cshl_fasta_formatter/1.0.1 |
| 1 | fastp | toolshed.g2.bx.psu.edu/repos/iuc/fastp/fastp/0.23.2+galaxy0 |
| 3 | VADR - Viral Annotation DefineR | testtoolshed.g2.bx.psu.edu/repos/f.touzain/vadr/vadr/0.1.0 |
| 4 | Map with BWA-MEM | toolshed.g2.bx.psu.edu/repos/devteam/bwa/bwa_mem/0.7.12.1 |
| 5 | bcftools mpileup | toolshed.g2.bx.psu.edu/repos/iuc/bcftools_mpileup/bcftools_mpileup/1.4.0.0 |
| 6 | VarDict on virus | toolshed.g2.bx.psu.edu/repos/iuc/vardict_java/vardict_java/1.8.3+galaxy1 |
| 7 | bcftools call | toolshed.g2.bx.psu.edu/repos/iuc/bcftools_call/bcftools_call/1.4.0 |
| 8 | vvv2_display: Display SNP proportions and CDS of an assembly in png image | testtoolshed.g2.bx.psu.edu/repos/f.touzain/vvv2_display/vvv2_display/0.1.10 |
| 9 | vcfutils_vcf2fq: convert vcf file to fastq file | testtoolshed.g2.bx.psu.edu/repos/f.touzain/vcfutils_vcf2fq/vcfutils_vcf2fq/1.16 |
| 10 | seqtk_seq fq2fa | fastq2fasta conversion toolshed.g2.bx.psu.edu/repos/iuc/seqtk/seqtk_seq/1.3.3 |
Outputs
| ID | Name | Description | Type |
|---|---|---|---|
| _anonymous_output_1 | _anonymous_output_1 | n/a |
|
| _anonymous_output_3 | _anonymous_output_3 | n/a |
|
| _anonymous_output_4 | _anonymous_output_4 | n/a |
|
| _anonymous_output_5 | _anonymous_output_5 | n/a |
|
| _anonymous_output_6 | _anonymous_output_6 | n/a |
|
| _anonymous_output_7 | _anonymous_output_7 | n/a |
|
| _anonymous_output_8 | _anonymous_output_8 | n/a |
|
| _anonymous_output_9 | _anonymous_output_9 | n/a |
|
| _anonymous_output_10 | _anonymous_output_10 | n/a |
|
Version History
Version 2 (earliest) Created 28th Jun 2023 at 10:52 by Fabrice Touzain
Update vadr wrapper (use --glsearch --split --cpu 4) and vadr version (1.5.1) to limit memory usage (especially for sarscov2 model)
Open
 master
master070a0c8
 Creators and Submitter
Creators and SubmitterCreator
Submitter
Activity
Views: 6693 Downloads: 1252 Runs: 6
Created: 28th Jun 2023 at 10:52
Last updated: 19th Jun 2025 at 11:23
Annotated Properties
Topic annotations
 Tags
Tags Attributions
AttributionsNone
 View on GitHub
View on GitHub Run on Galaxy
Run on Galaxy https://orcid.org/0000-0002-1103-2425
https://orcid.org/0000-0002-1103-2425